Introduction of Help
FGD is a comprehensive genome and transcriptome database for multi-omics research of Ficus species, FGD includes:
-Genome assemblies and annotations of five inbred Ficus species: Ficus carica, Ficus erecta, Ficus microcarpa ,Ficus hispida and Ficus pumila;
-Transcriptome raw and assembled sequence , annotations of 24 Ficus fig ostiolet bracts CDS sequence;
-Transcriptome expression data of three ficus bracts with reference genomes(.bam file Format), the three Ficus species are Ficus erecta, Ficus hispida, and Ficus microcarpa;
-Expression patterns of tissues from different development stage of same inbred line and same tissue of different samples;
-Genomic collinearity data and graphical presentation of different Ficus species;
-ncRNA information of five Ficus species, It includes SSR and tRNA information for each genome and is displayed in the GBrowse browser;
-Intergrated Terpene Synthase(TPS) Genes mining tools in FGD, this tookit can identify TPS genes from protein sequences and files;
-Integrated primer3 primer design function and GO enrichment analysis function.
In short,FGD generated comprehensive functional annotations for the annotated gene models in each assembly, and provided useful tools for users to search, analyze and visualize all these different omics data.
Sequence Search
Step 1 Select Species to show

Step 2 Select one type
Step 3 Select the source of the corresponding species
Step 4 Search for sequences by the location
Step 5 Search for sequences by entering name
Step 6 Click "Search" button to search
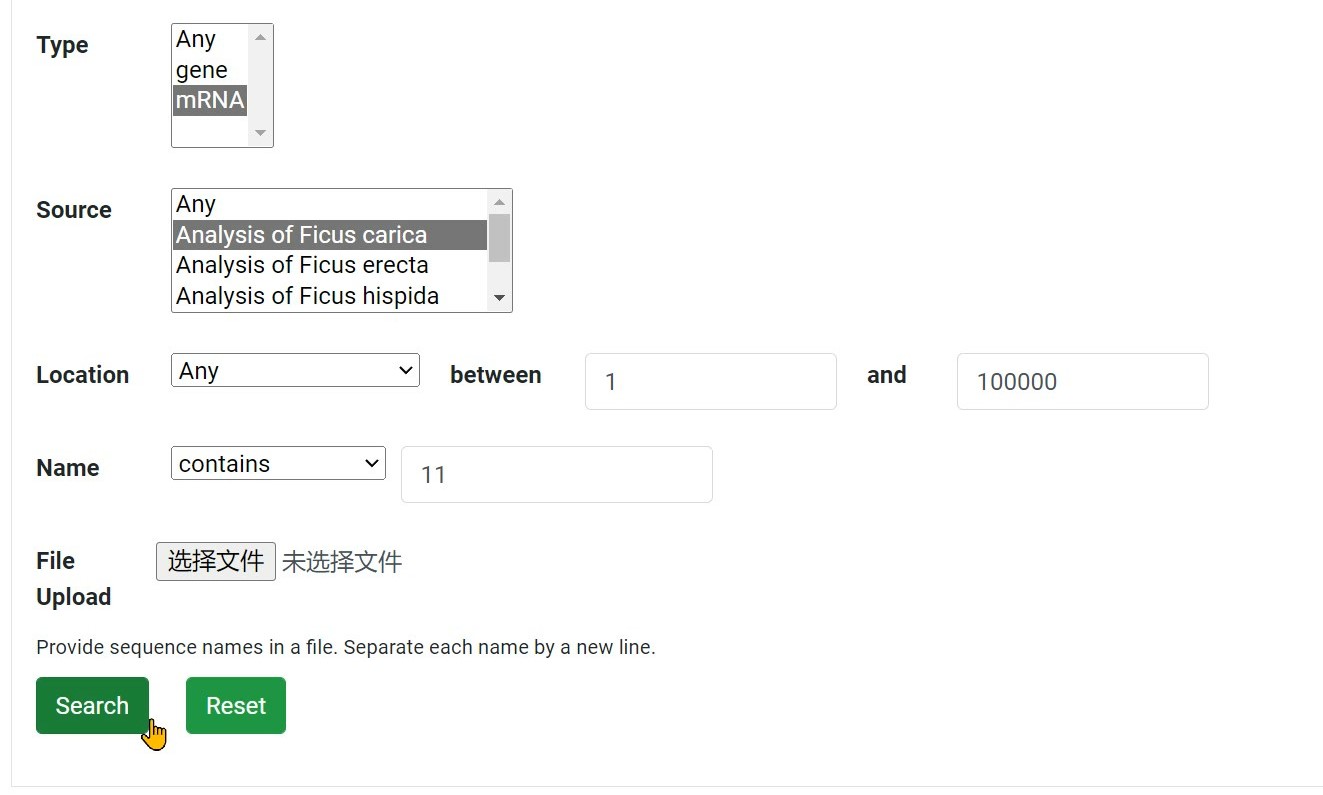
Step 7 Get the result
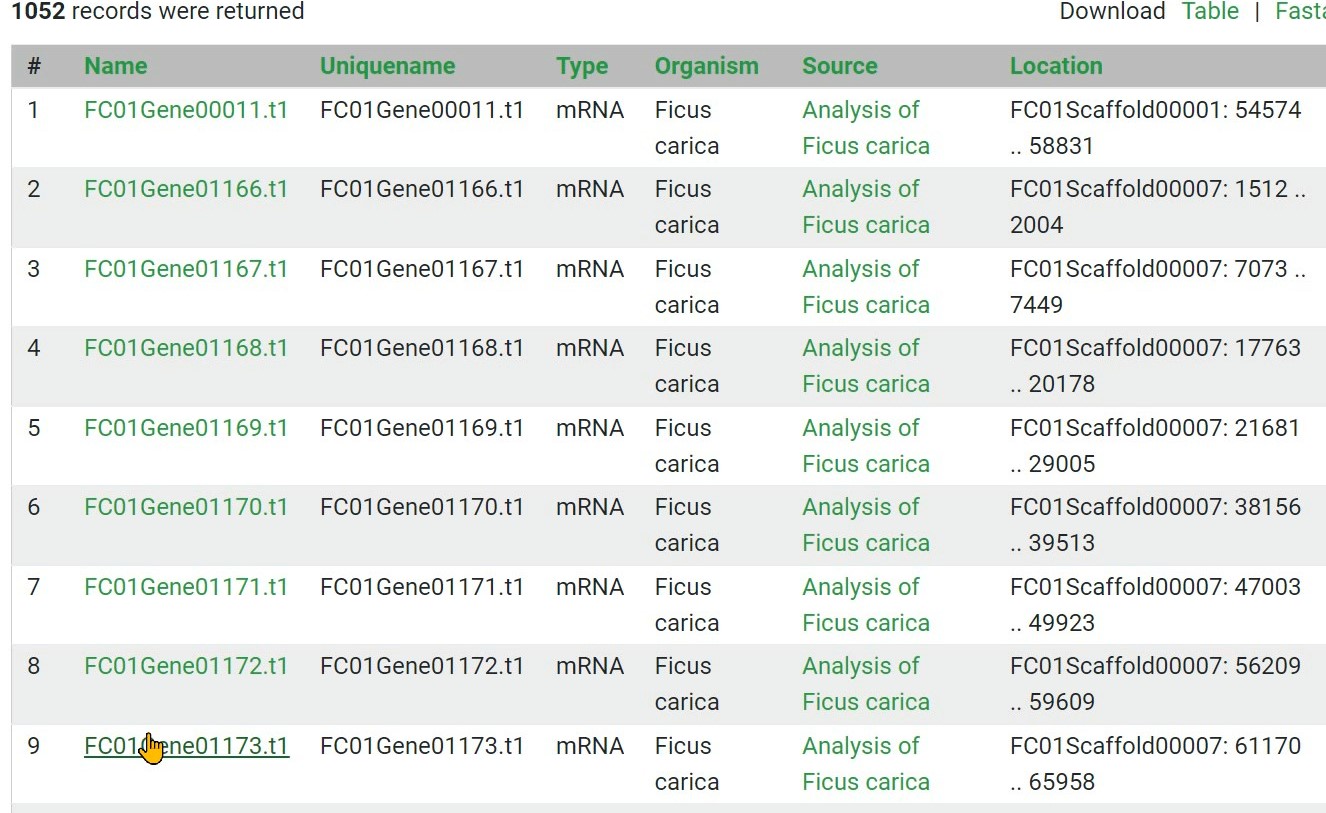
Step 8 The results can be downloaded in FASTA or CSV tabular format
Gene & Analysis search
Step 1 In the "Gene Search" Section, You Can Use It in Many Ways, Such as:
1) Identifier
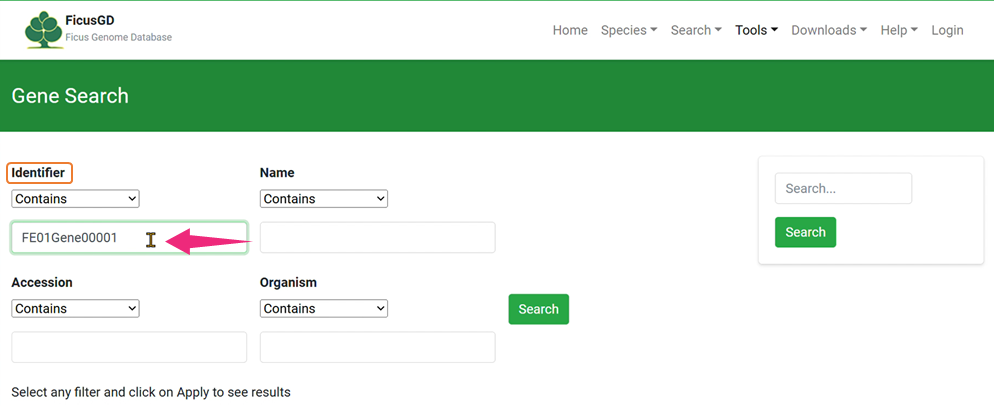
2)Organism
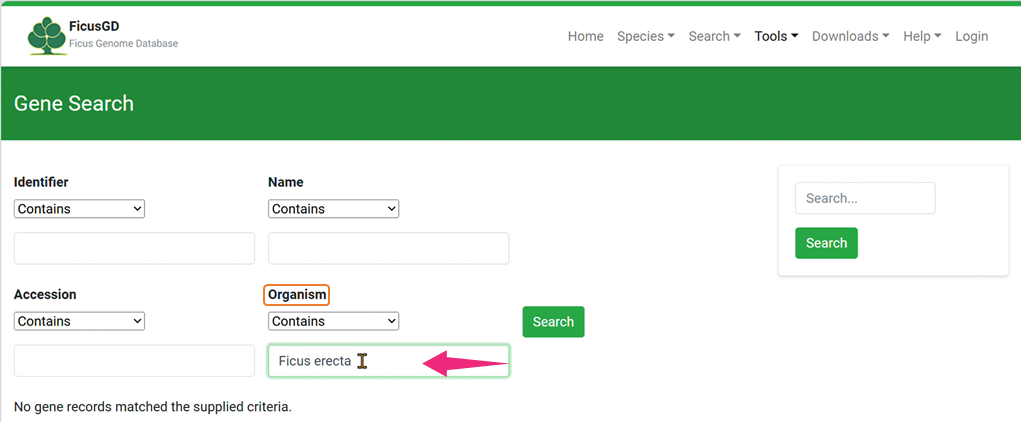
Click the "Search" Button to Get Your Result 3) Example of “Identifier”
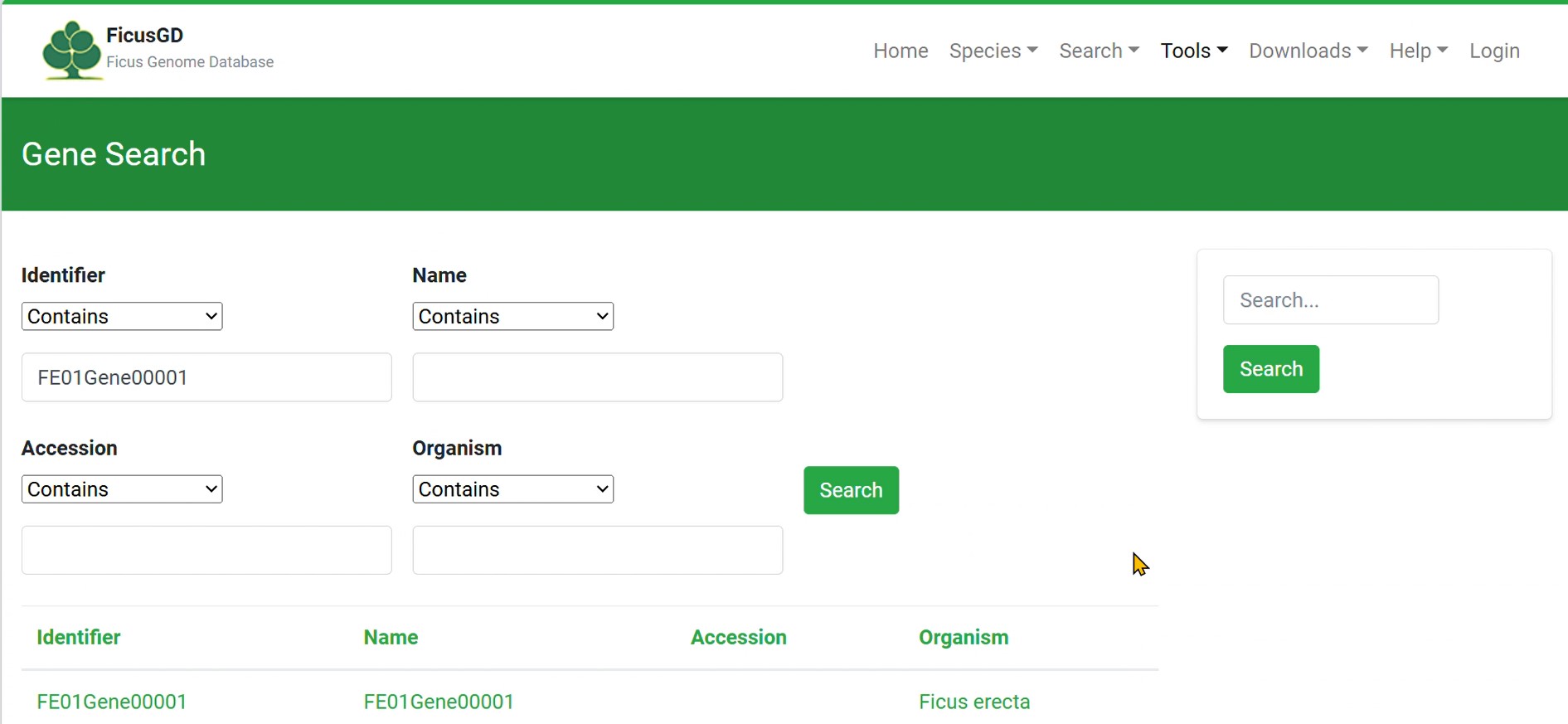
4) Example of “Organism”
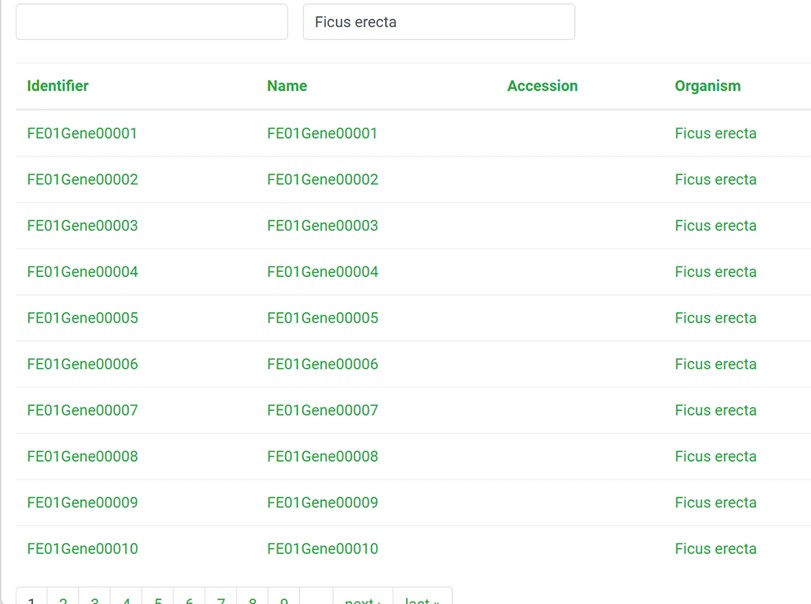
Step 2 In the "Analysis Search" Section, Input Keywords for Search
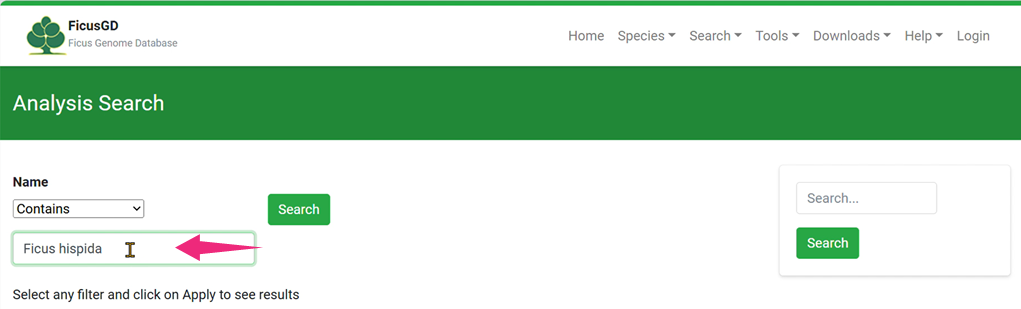
MegaSearch
Step 1 Select Data Type to show

Step 2 Select a field
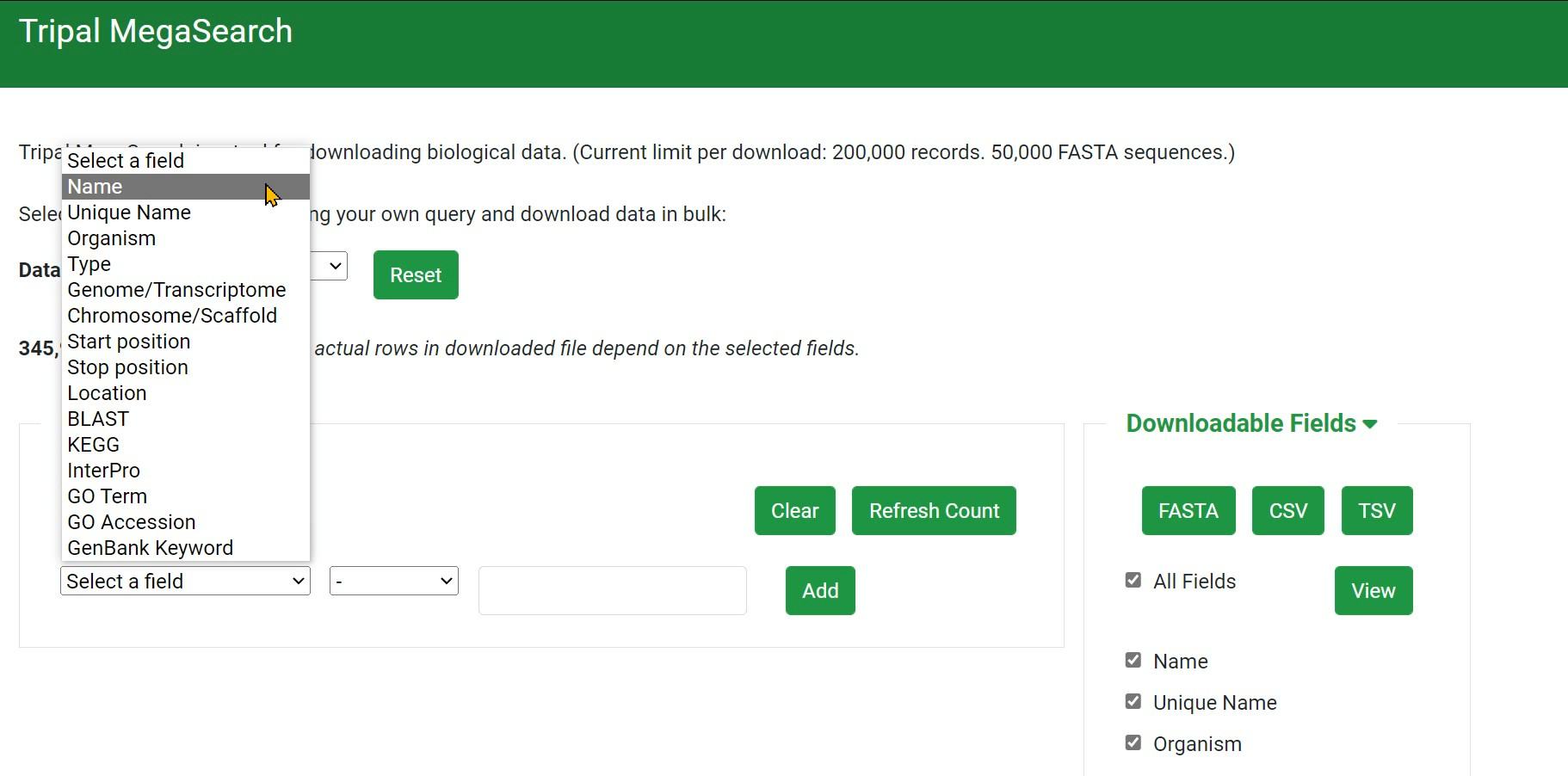
Step 3 Build your own query

Step 4 Download data in bulk: choose FASTA, CSV or TSV

Step 5 You can view the data online instead of downloading it
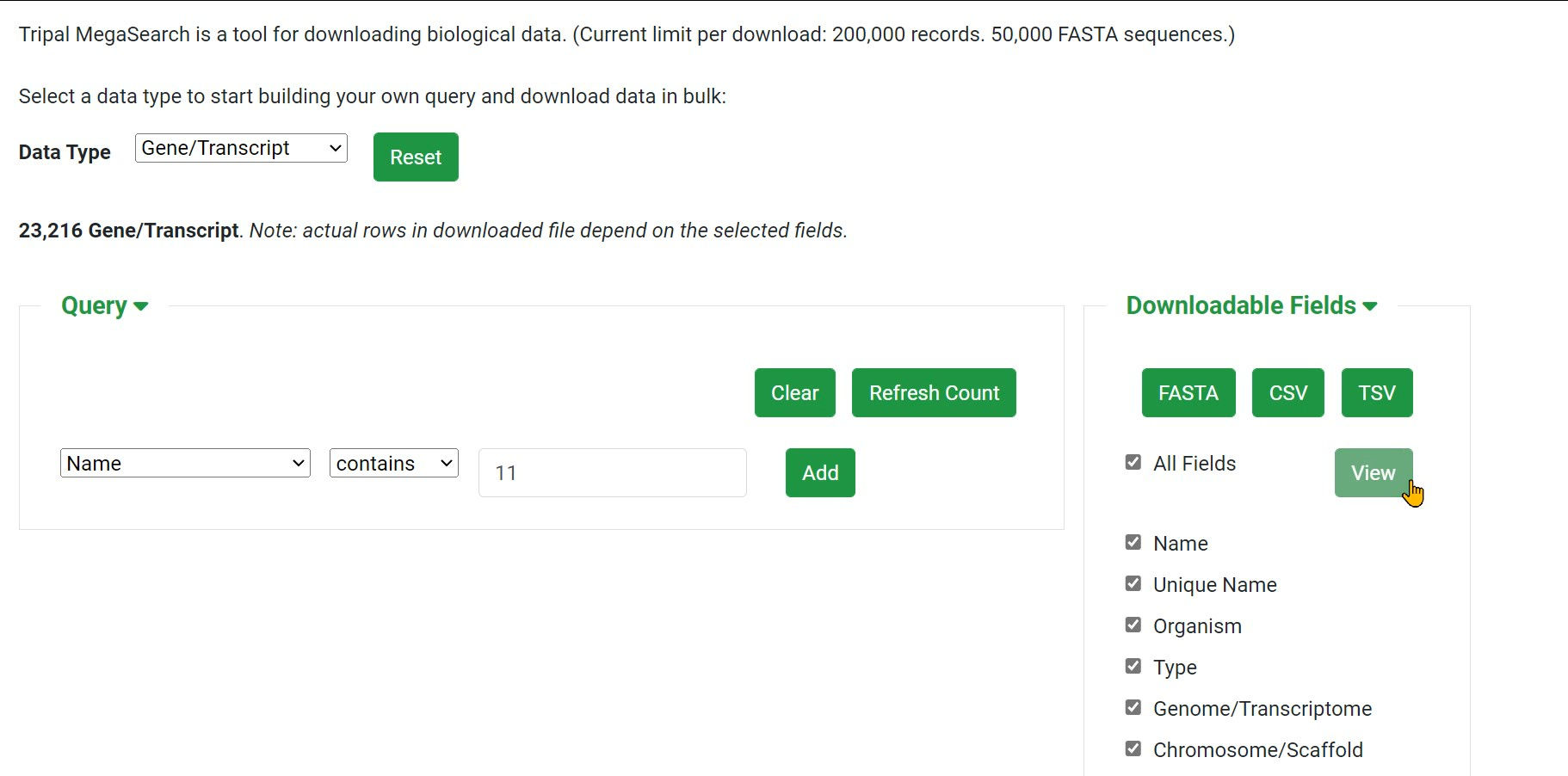
Step 6 The downloaded data contains more annotation
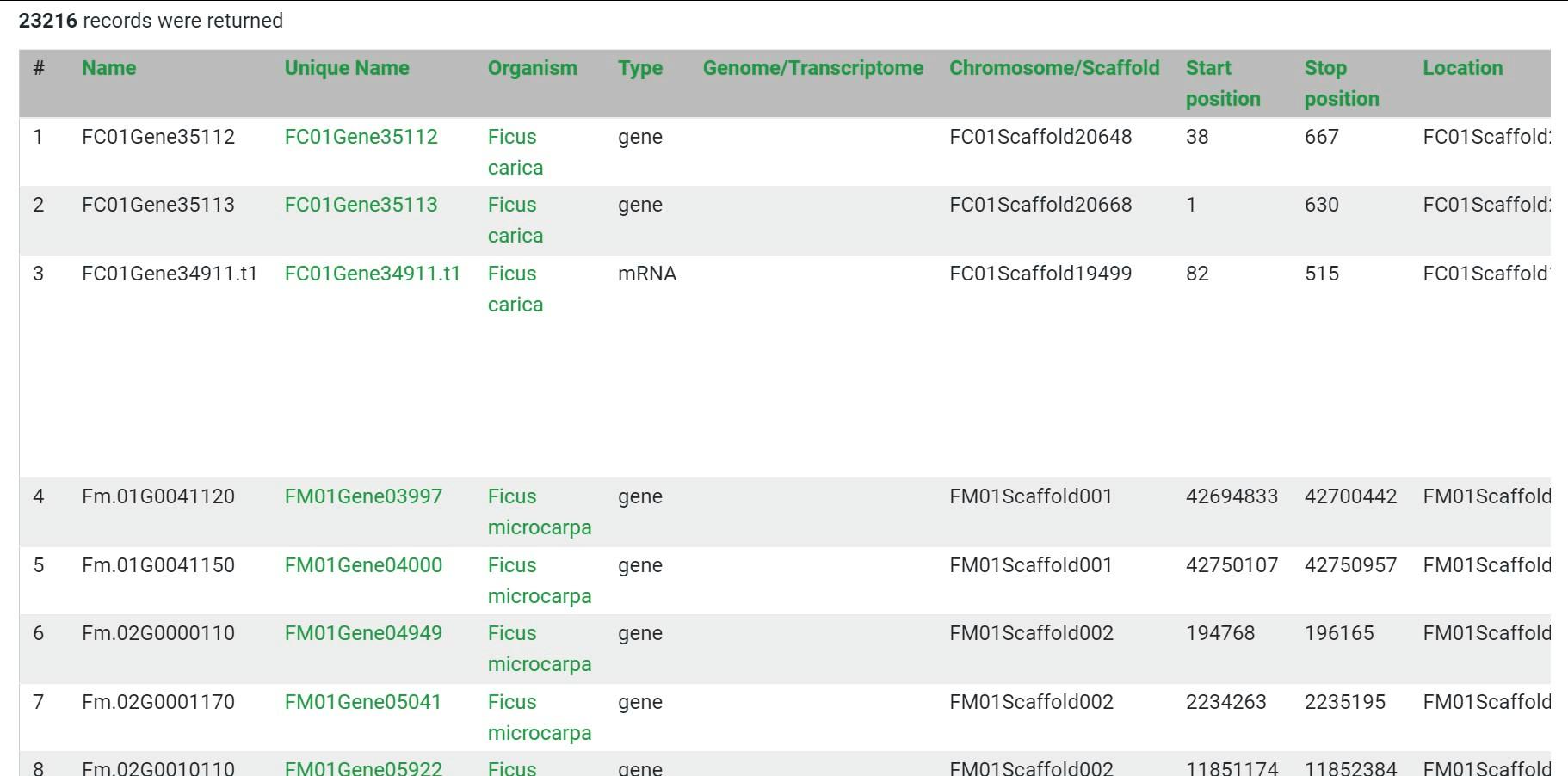
SyntenyViewer
Step 1 Select "Tools" and click " Synteny Viewer" button
Step 2 Select genome to show
Step 3 Select a chromosome/scaffold
Step 4 Choose (a) genome(s) for comparison
or
Step 5 Click "Search" button to search
Step 6 Get the synteny viewer provided in the circular view
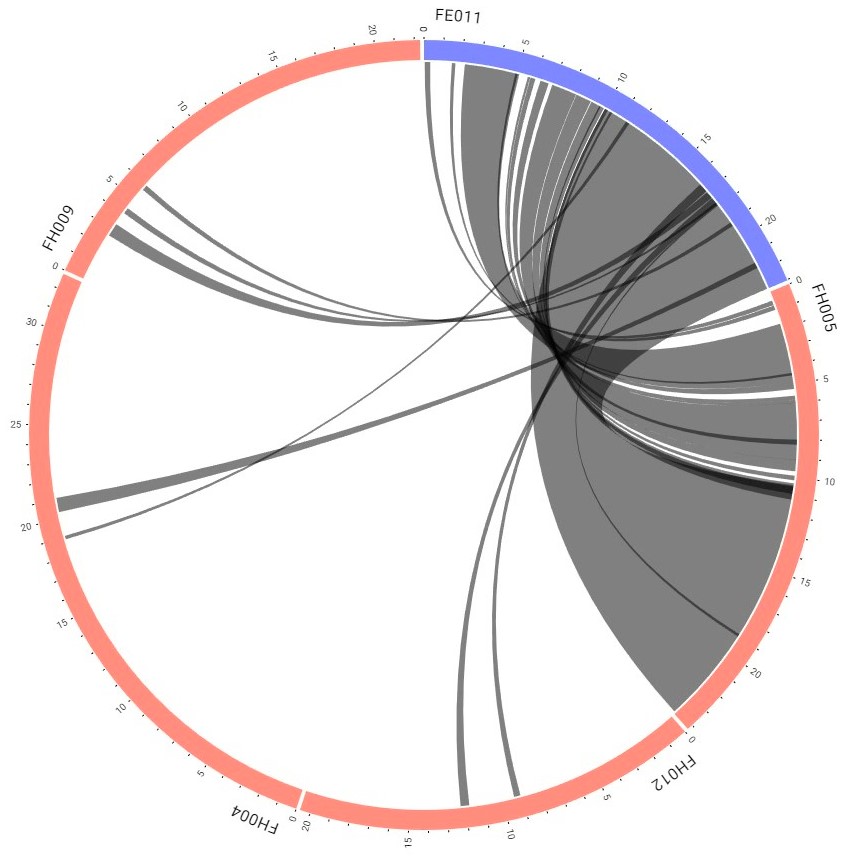
Step 7 Click the interested one to leads you to the detailed block information
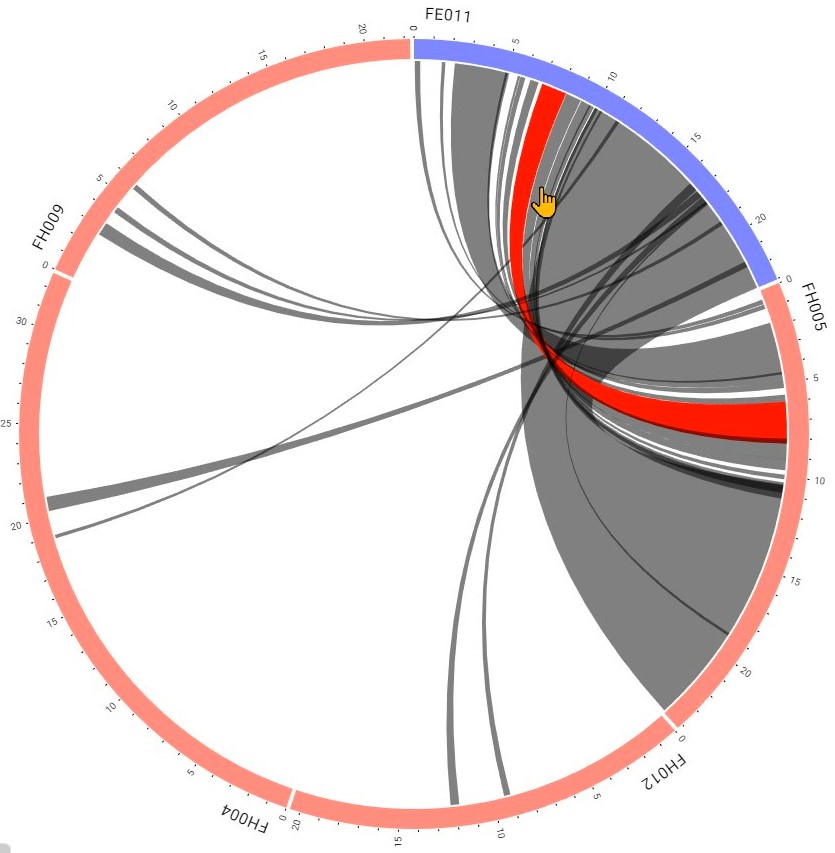
Step 8 Get the detailed information: the block information & the visualization
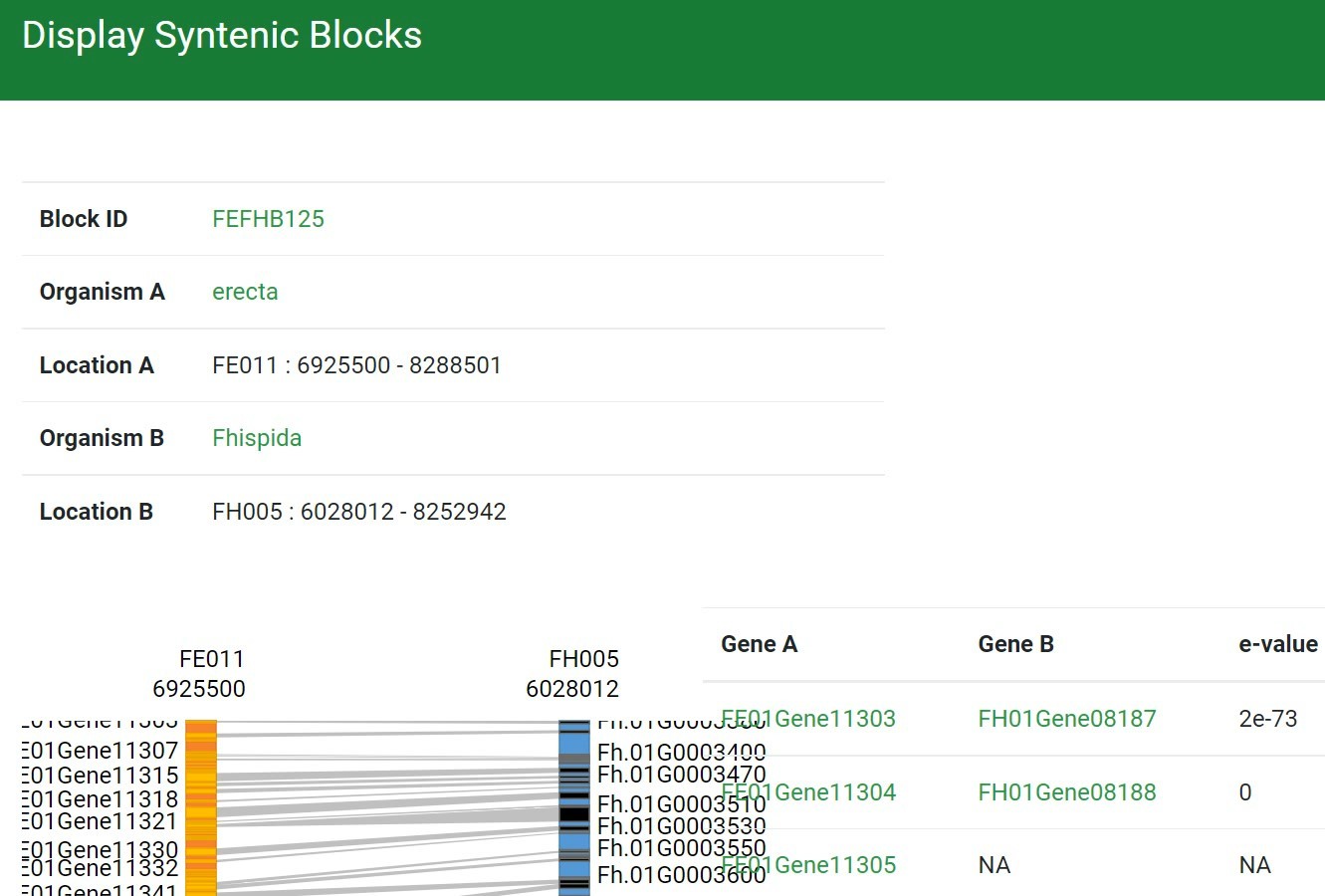
Step 9 Click the interested one
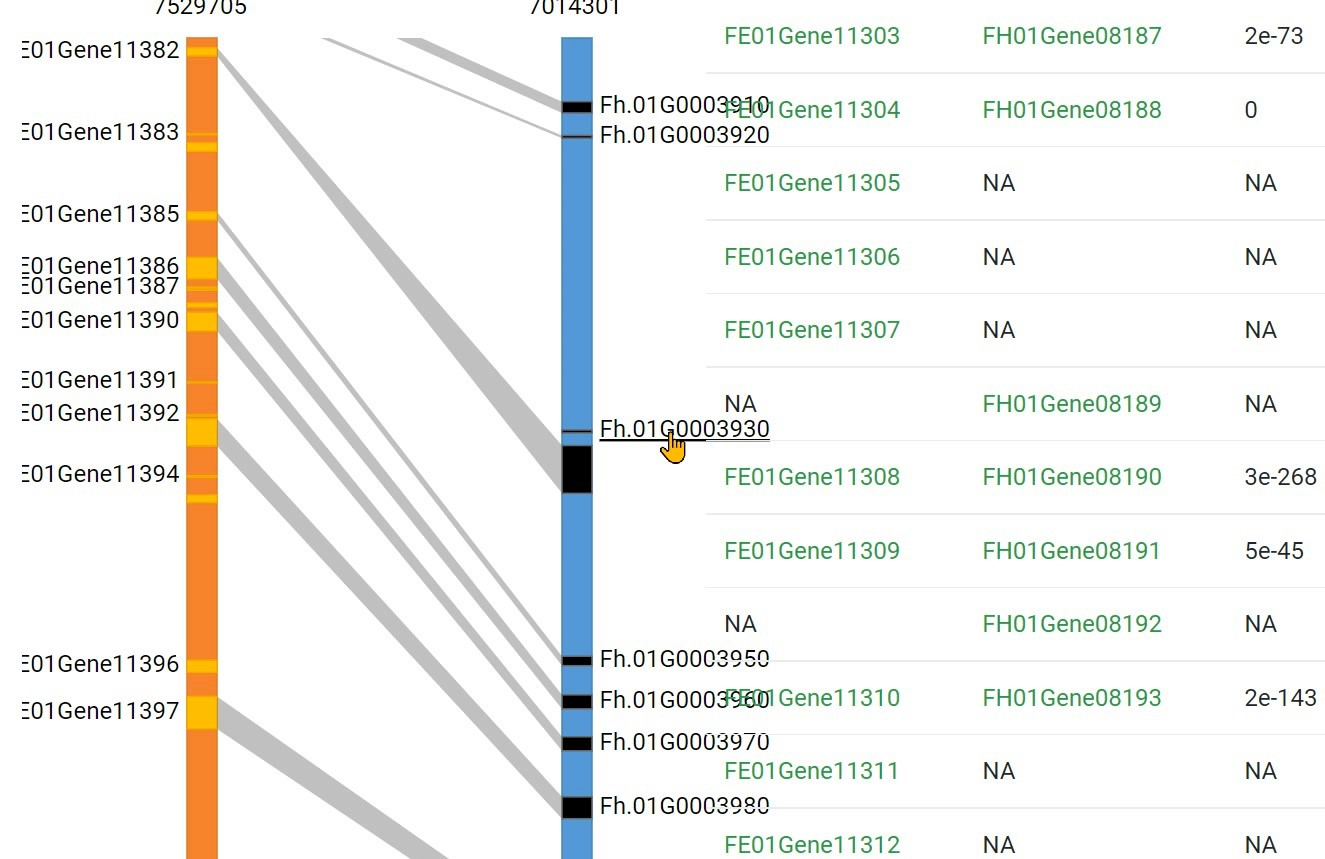
Step 10 Get the Result
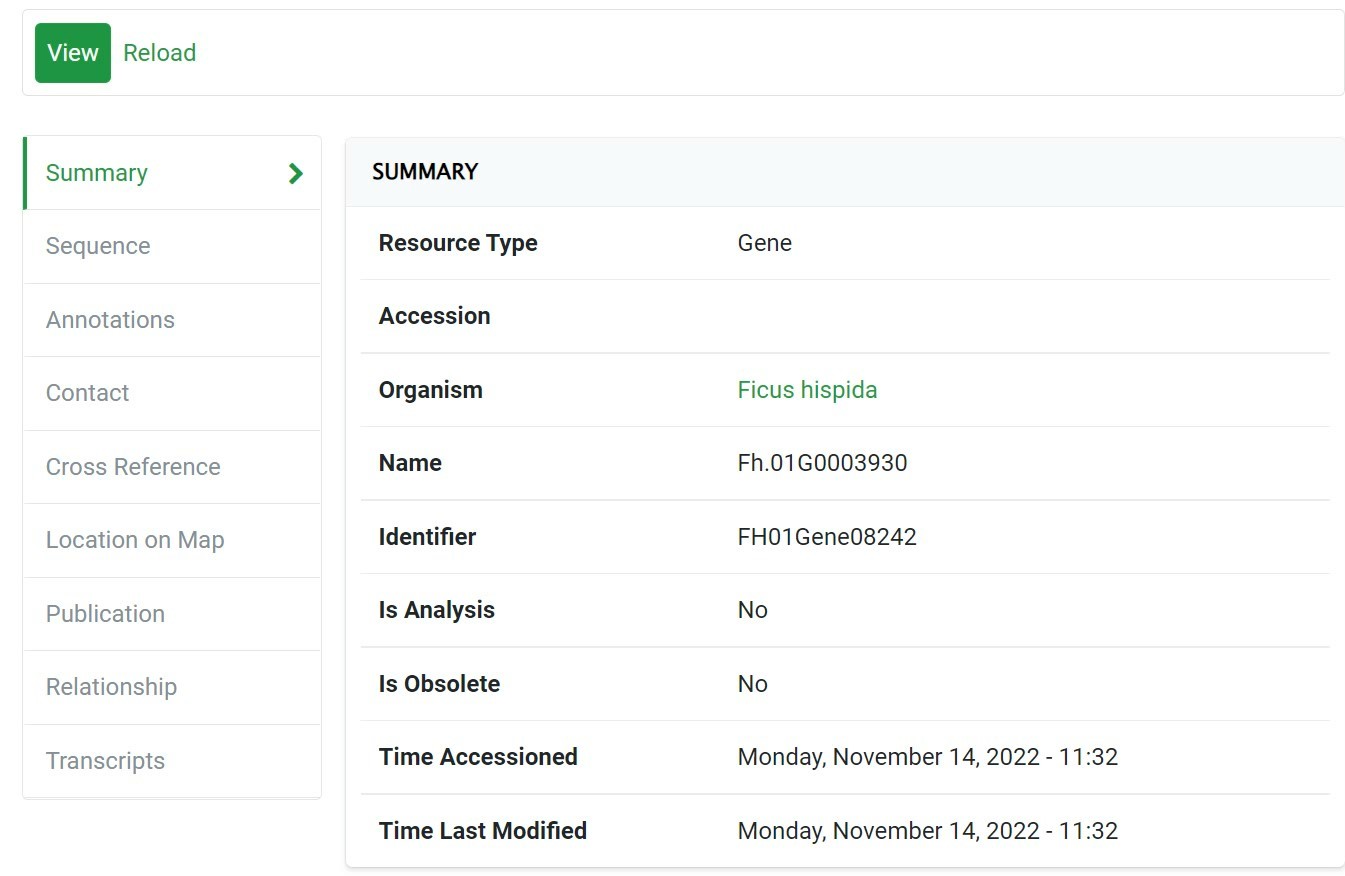
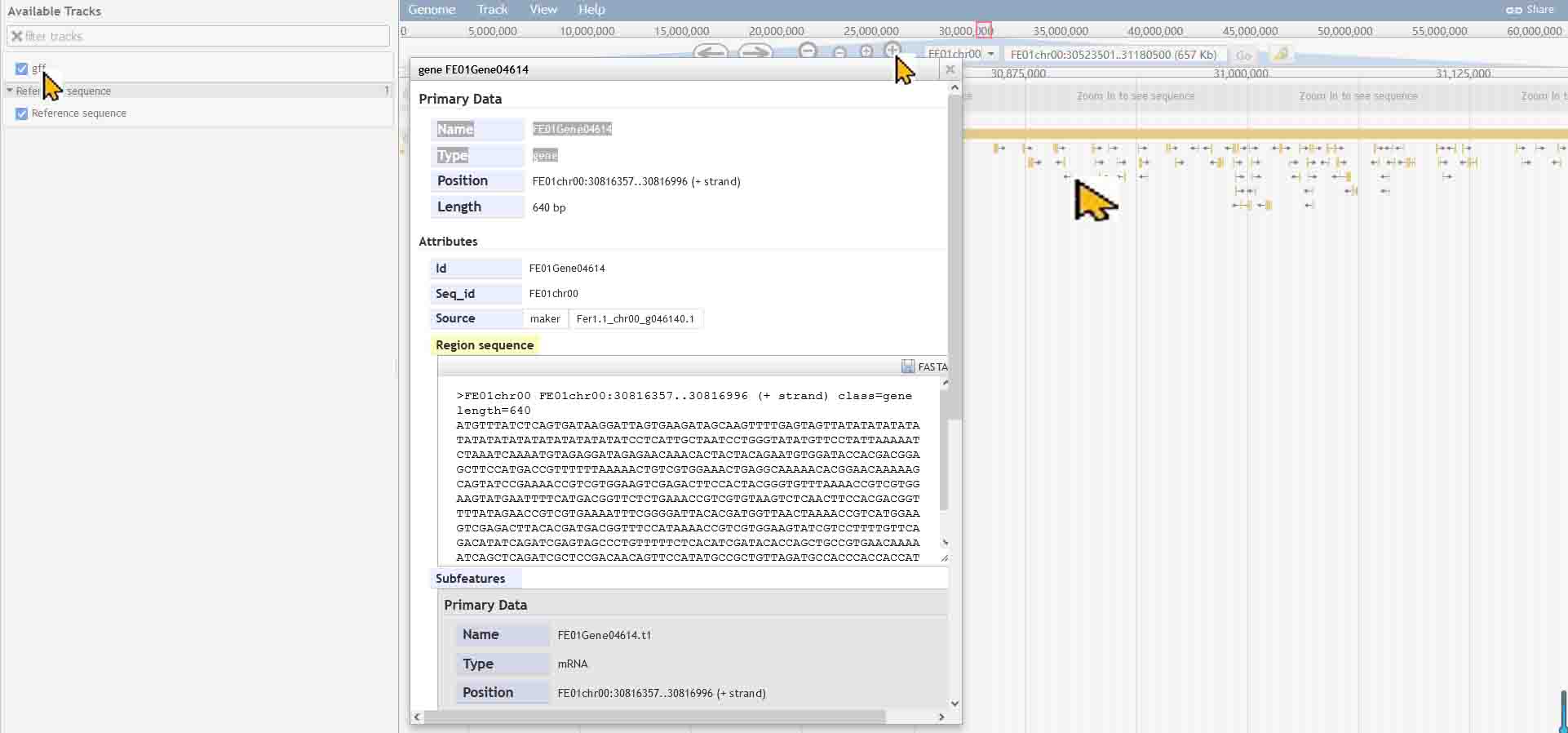
JBrowse
Usually,click on 'gff','+' ,'→' as the picture below, you would see the detailed information of every gene.
For more detailed help,please click on this HYPERLINK of YOUTUBE!
GBrowse
GBrowse
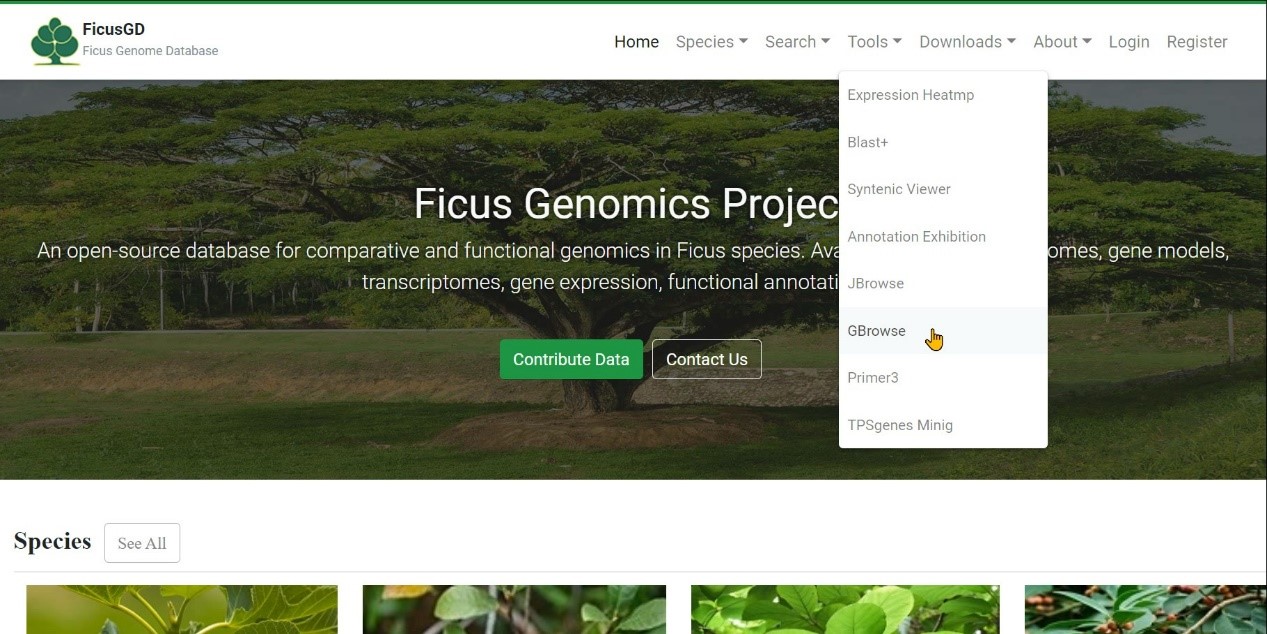
Step 1 Select "Tools" button
Step 2 Click "GBrowse" button
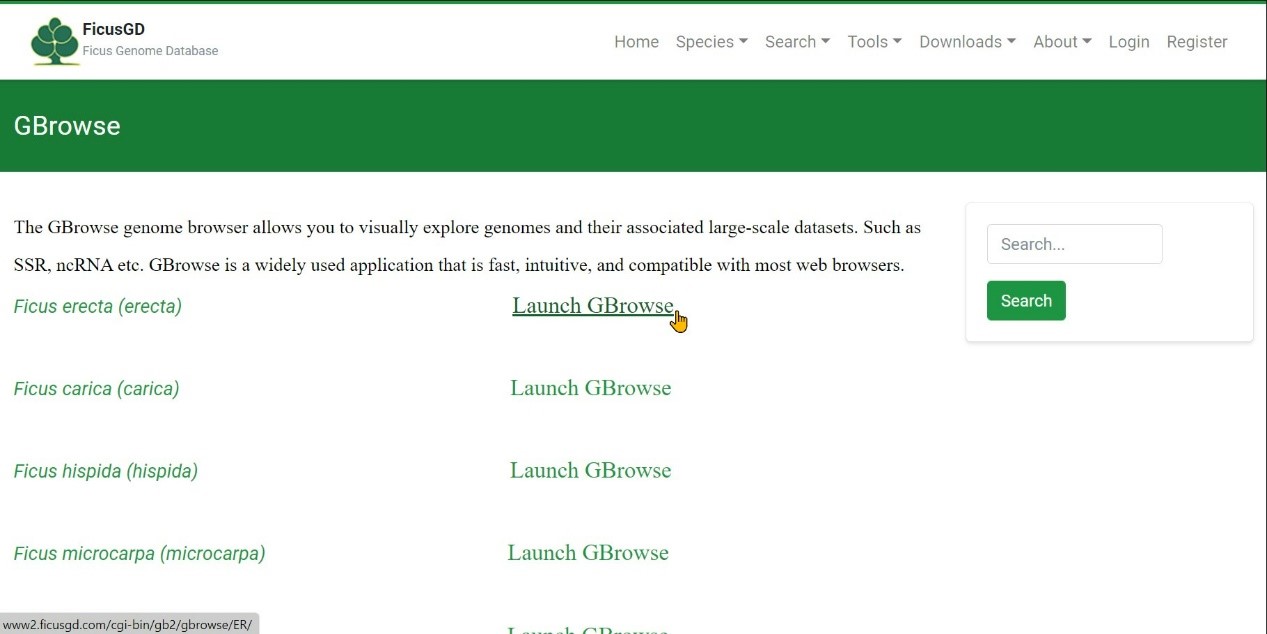
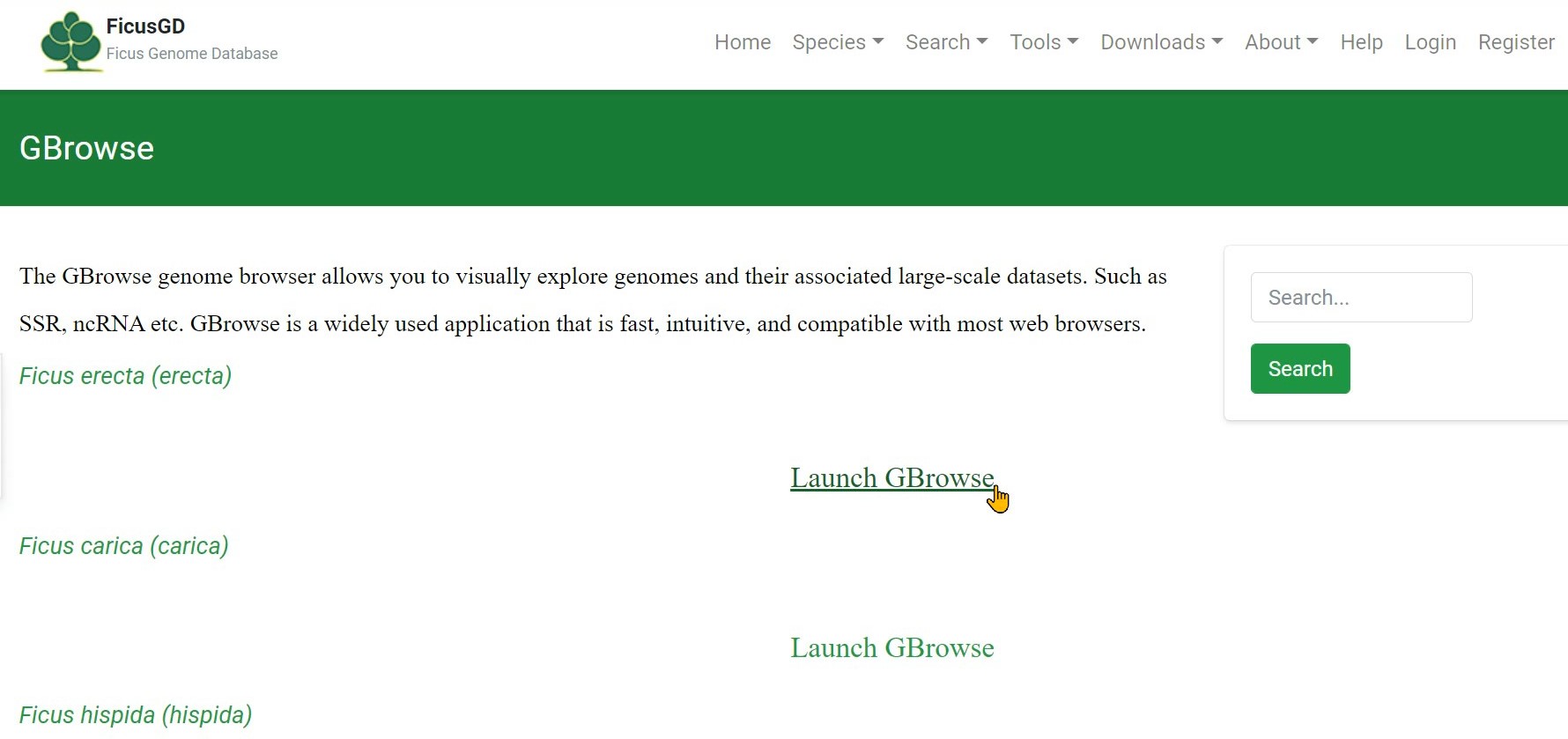
Step 3 Select one species and click "Launch GBrowse" button
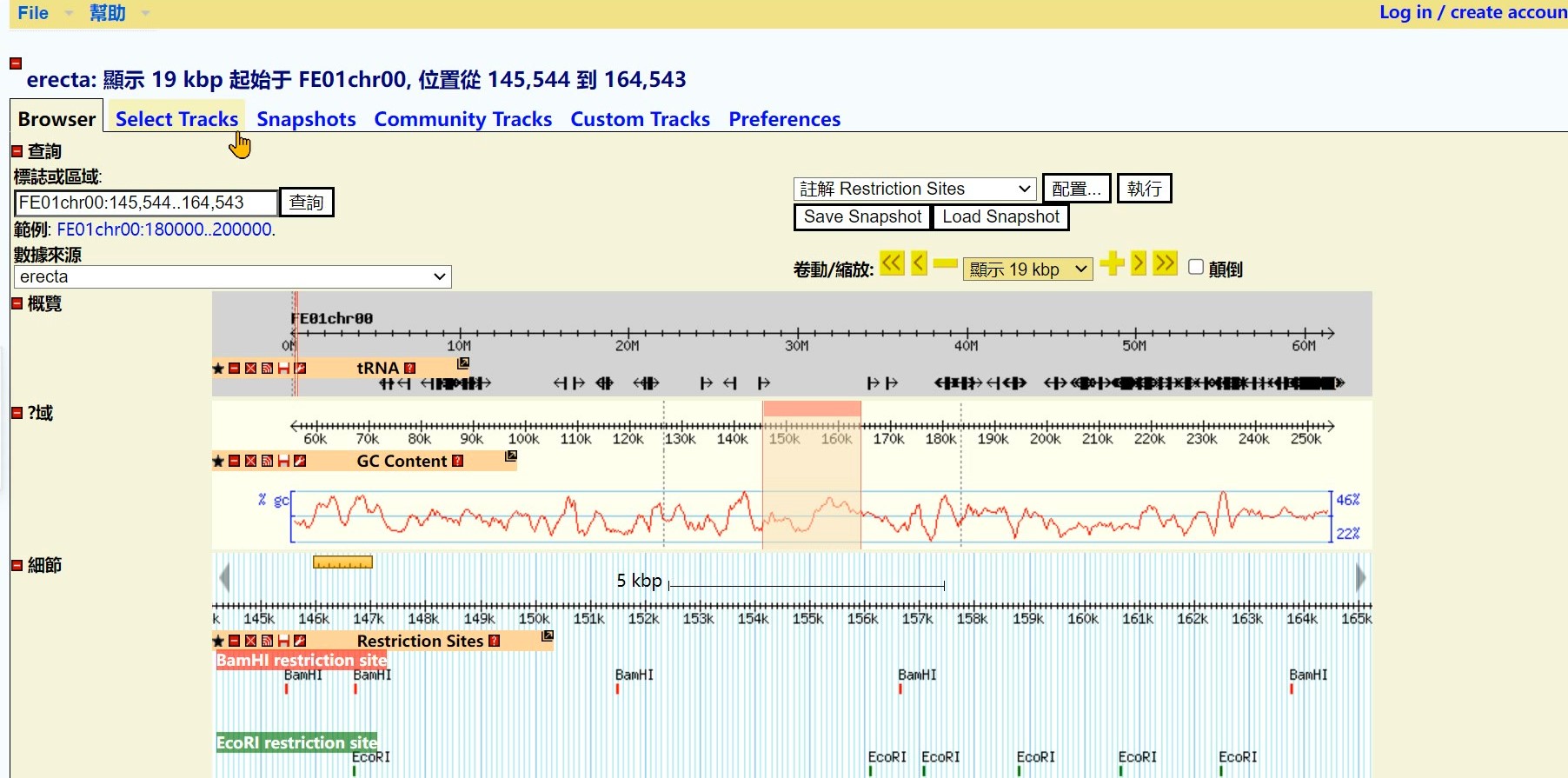
Step 4 Click "Select Tracks" to search the gene
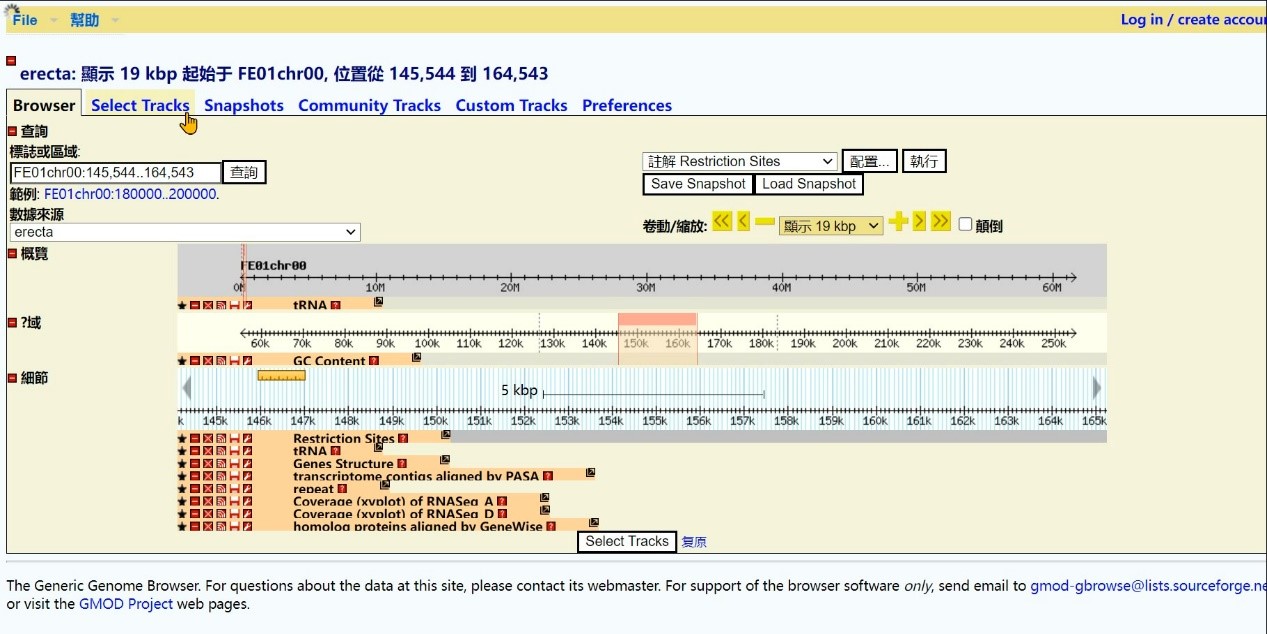
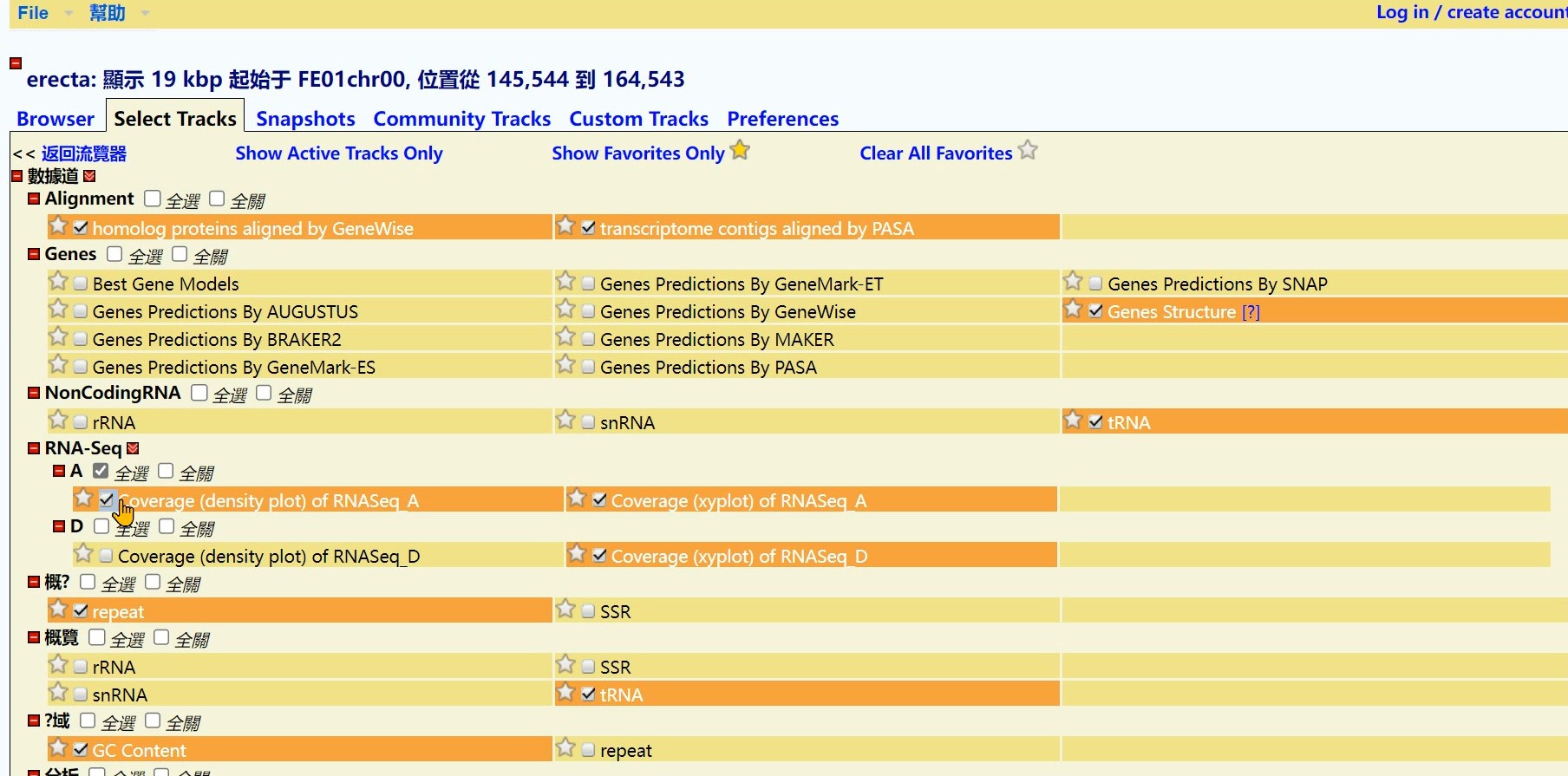
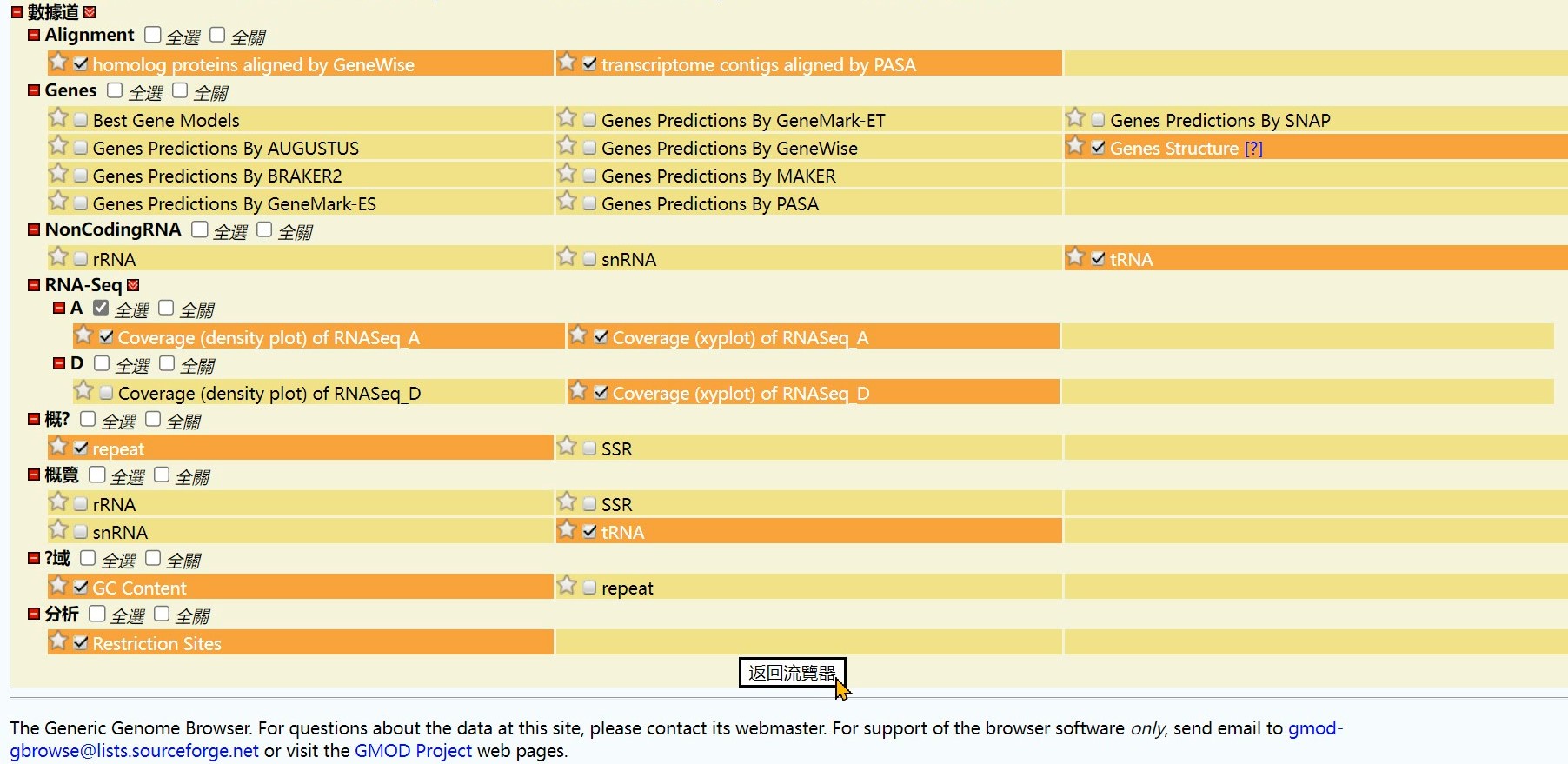
Step 5 Select search type, Click "tRNA"
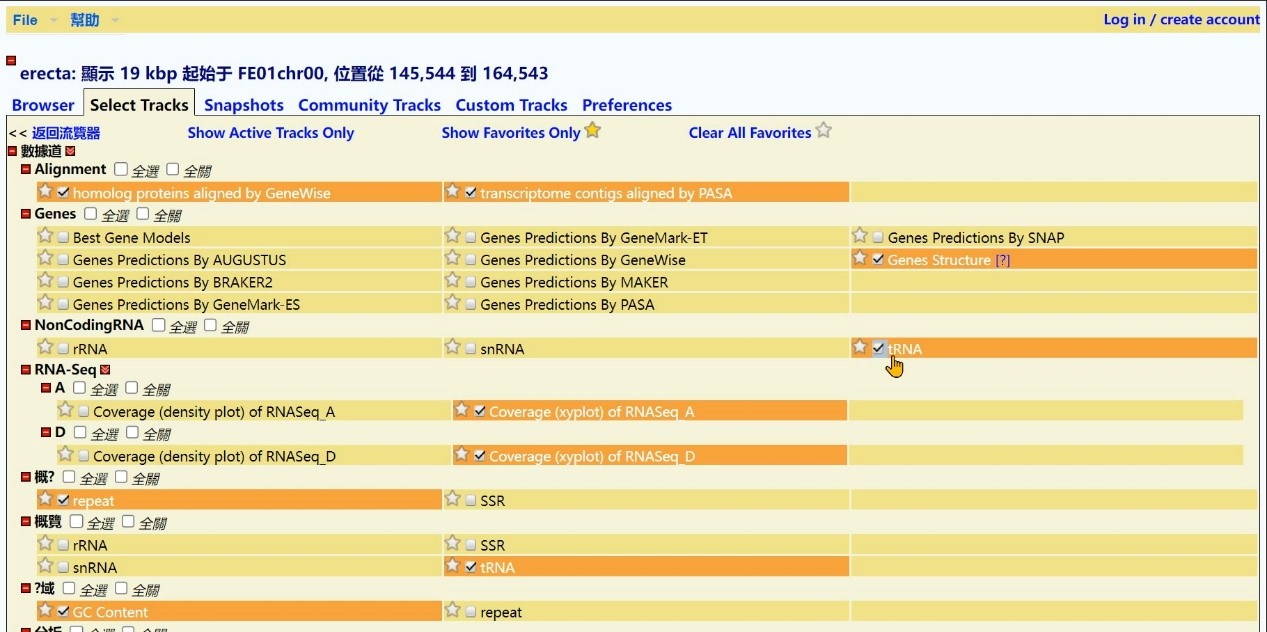
Step 6 Click the “Select All” button
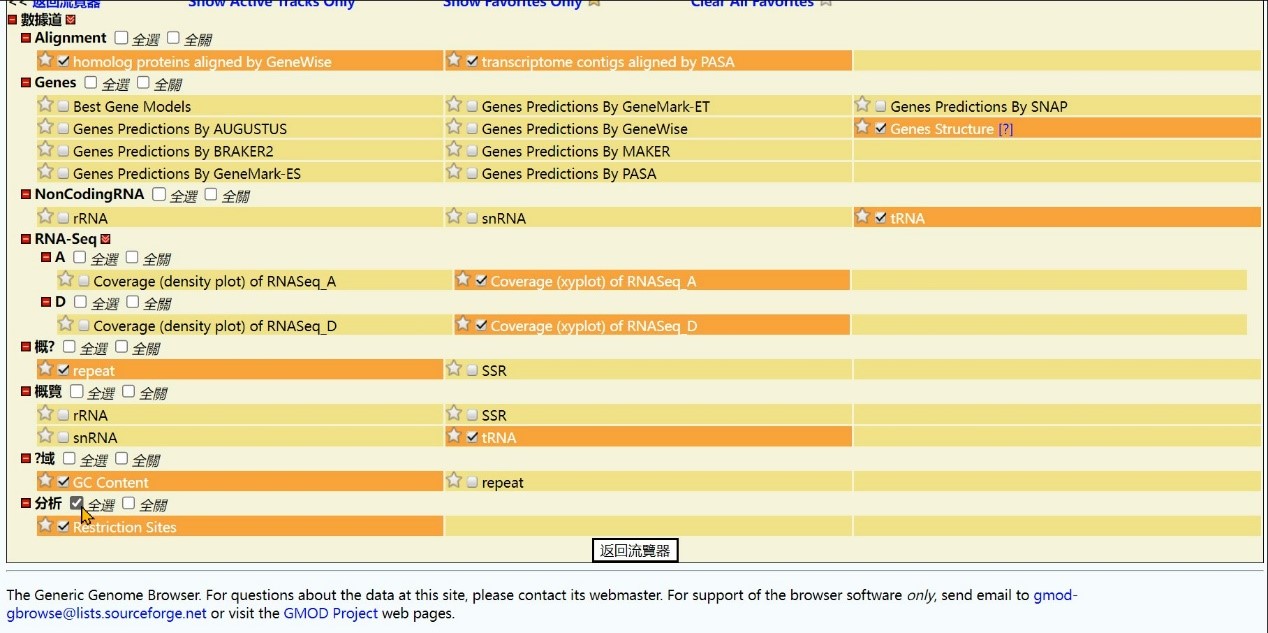
Step 7 Click the “Back to Browser” button
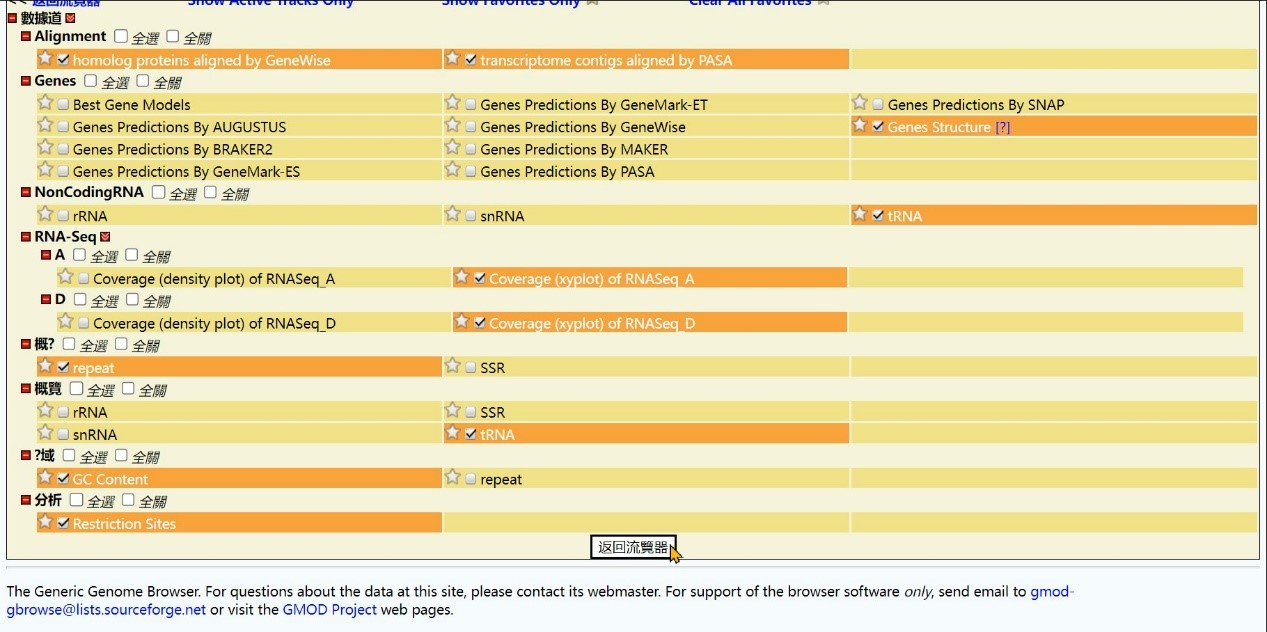
Step 8 Get the Result
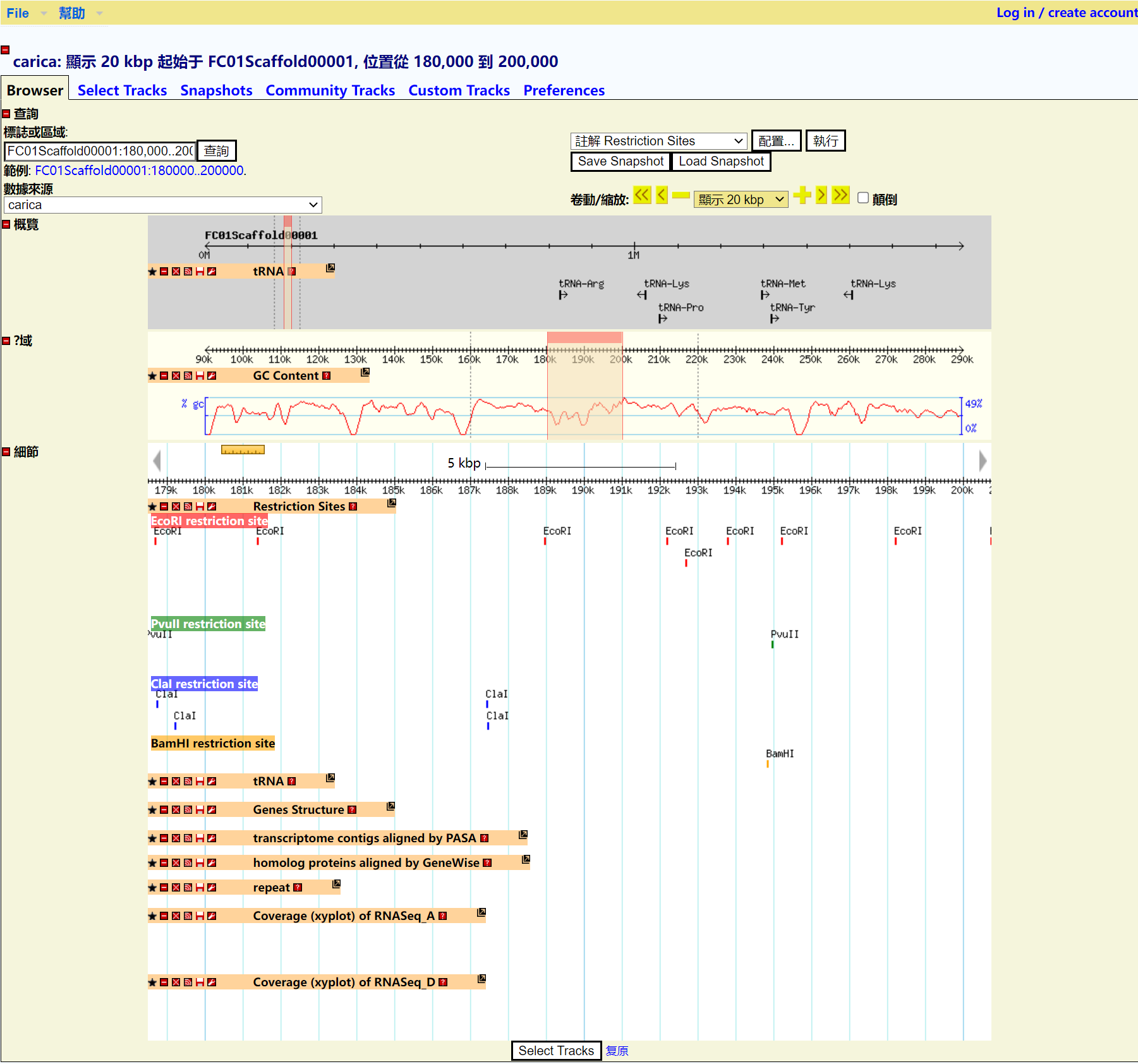
RNA-seq of GBrowse
Step 1 Click "Launch GBrowse" button
Step 2 Click "Select Tracks" to search the gene
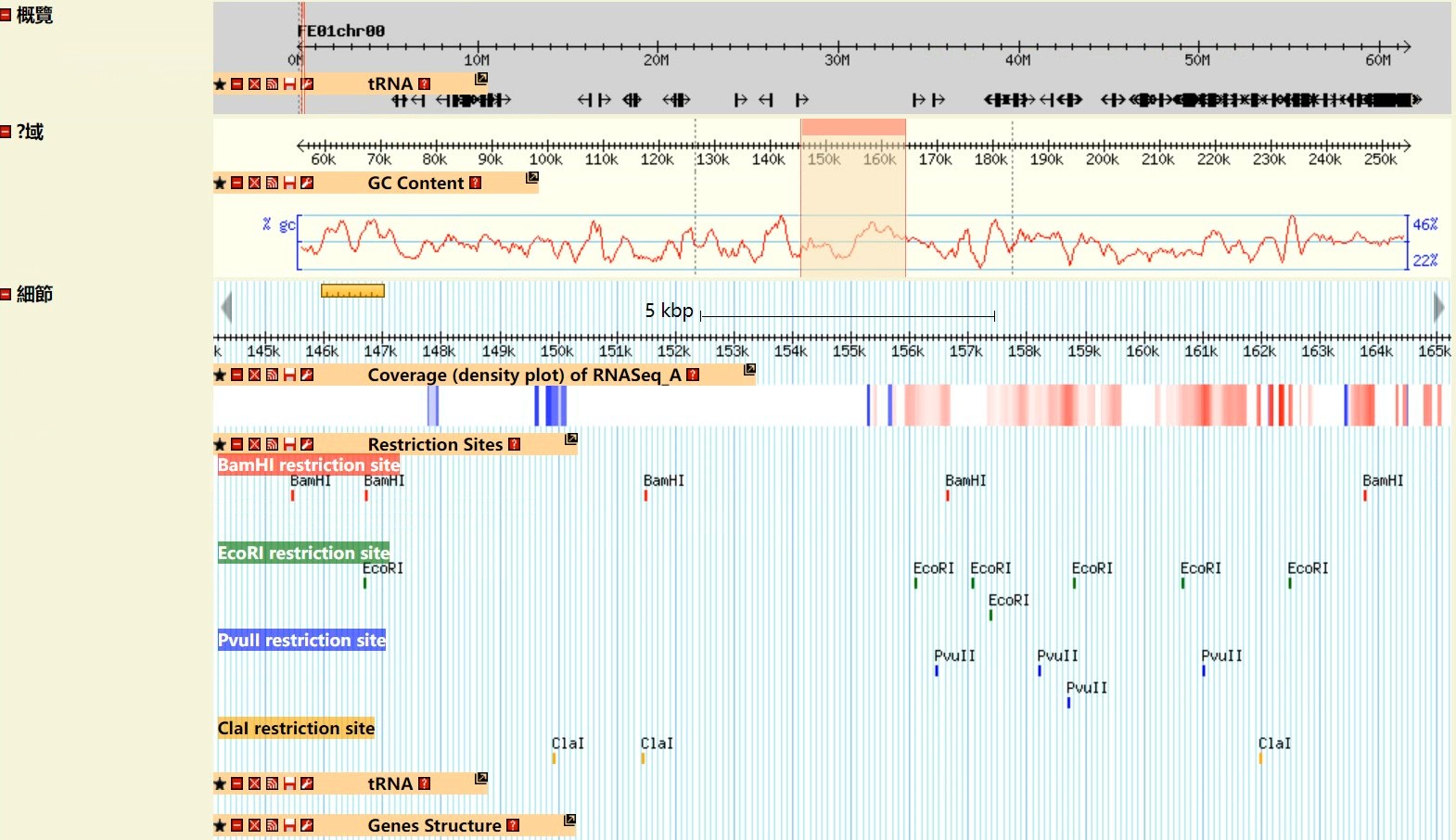
Step 3 Click the “Coverage (density plot) of RNASeq_A” button
Step 4 Click the “Back to Browser” button
Step 5 Get the Result
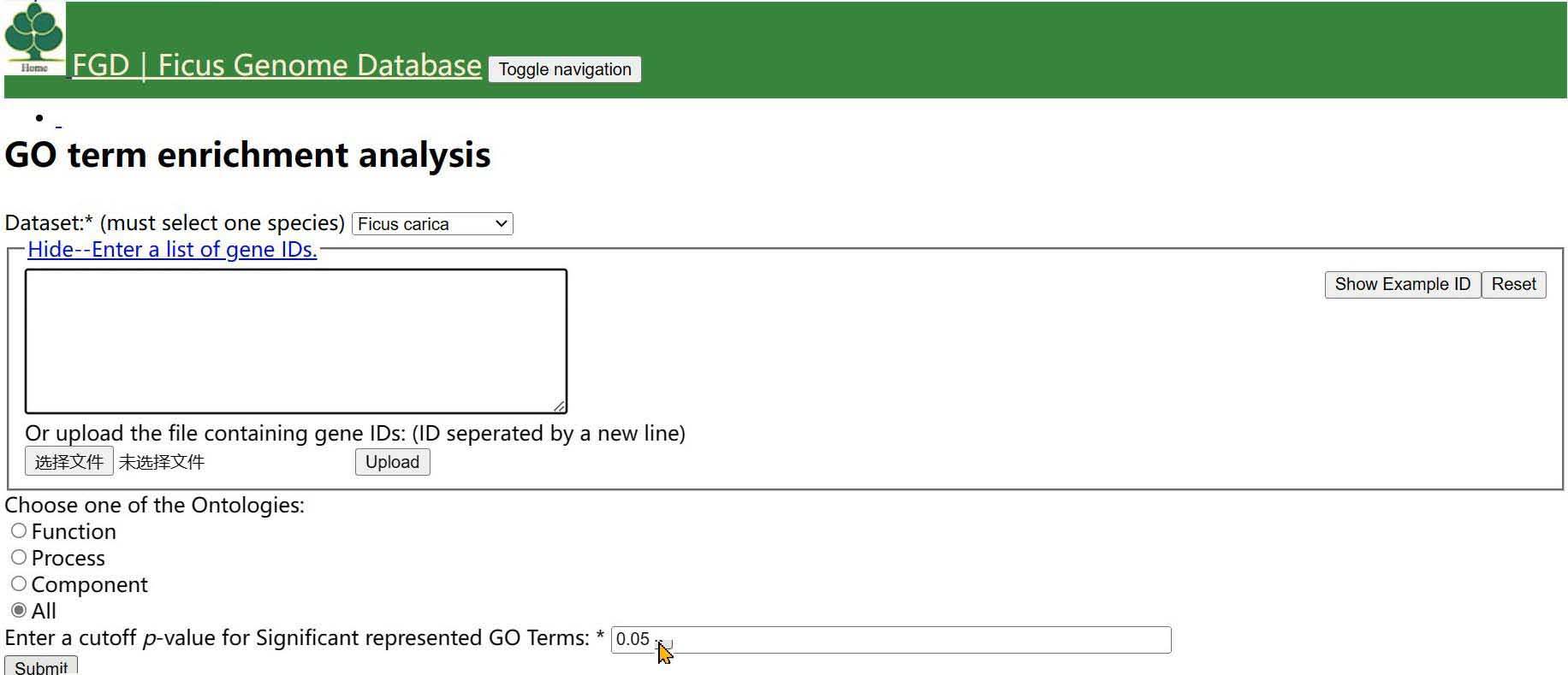
GO enrichment
Step 1 Select one species and click "Select a Dataset " button
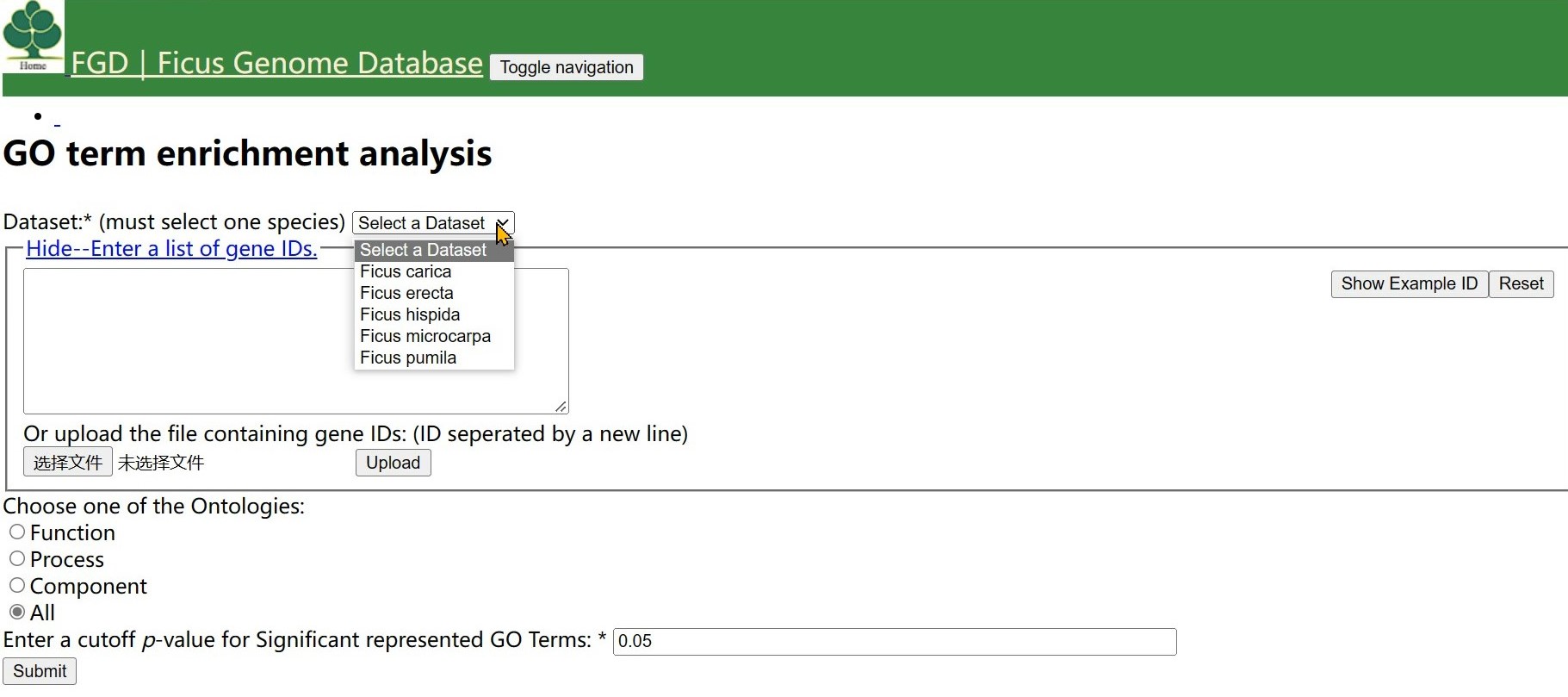
Step 2 Enter a list of gene IDs or upload the file containing gene IDs.
Tips:The gene IDs must conform to the style "FC01Gene00000",
the second alphabet can be H(hispida)E(erecta)M(microcarpa)P(pumila),
the last five letter must be Arabic numbers.Such as FE01Gene00234.
The detailed genes information can be downloaded from http://www.ficusgd.com/node/3.

or upload the file containing gene IDs(cannot enter and upload simultaneously)

Step 3 Choose one of the Ontologies and click the button
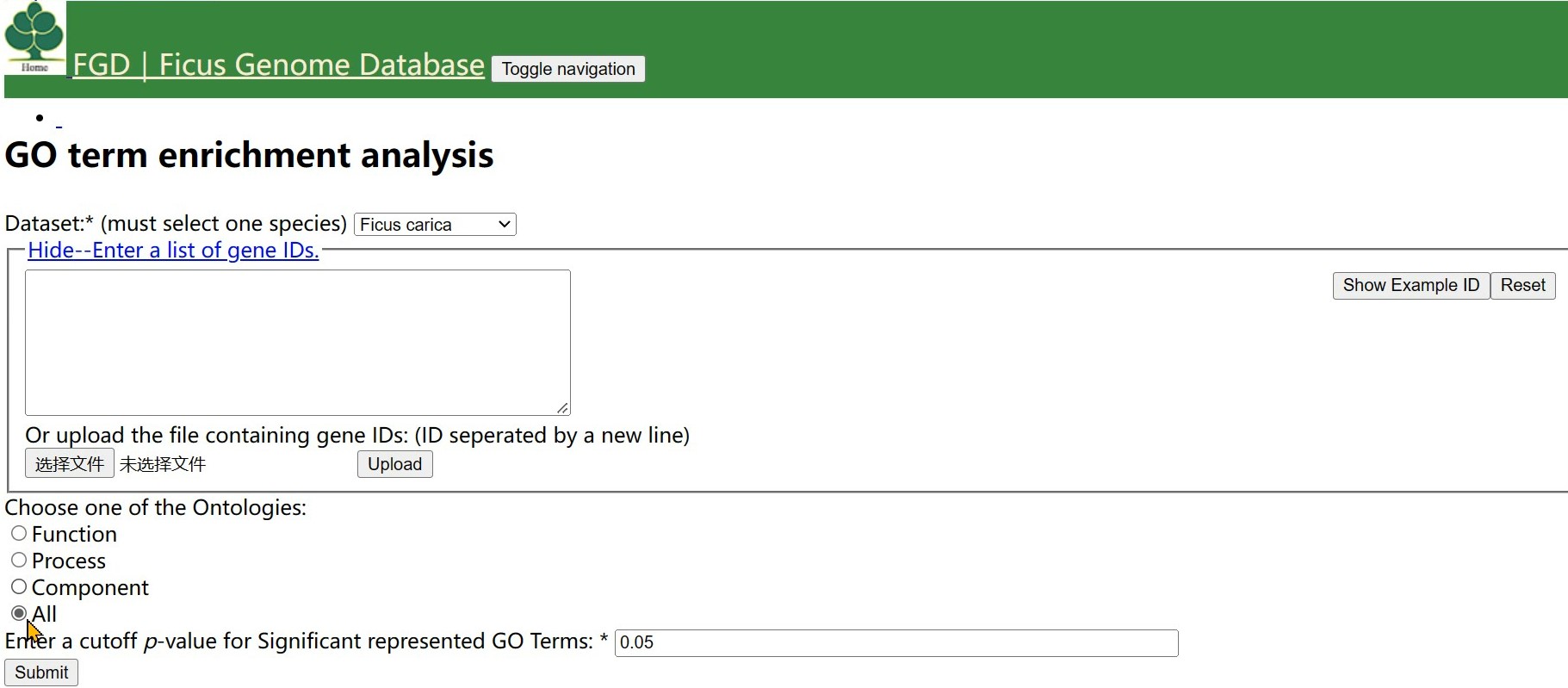
Step 4 Input a cut-off pavlue
Step 5 You can choose one of correction method,(Bonferroni correction is the most rigorous)
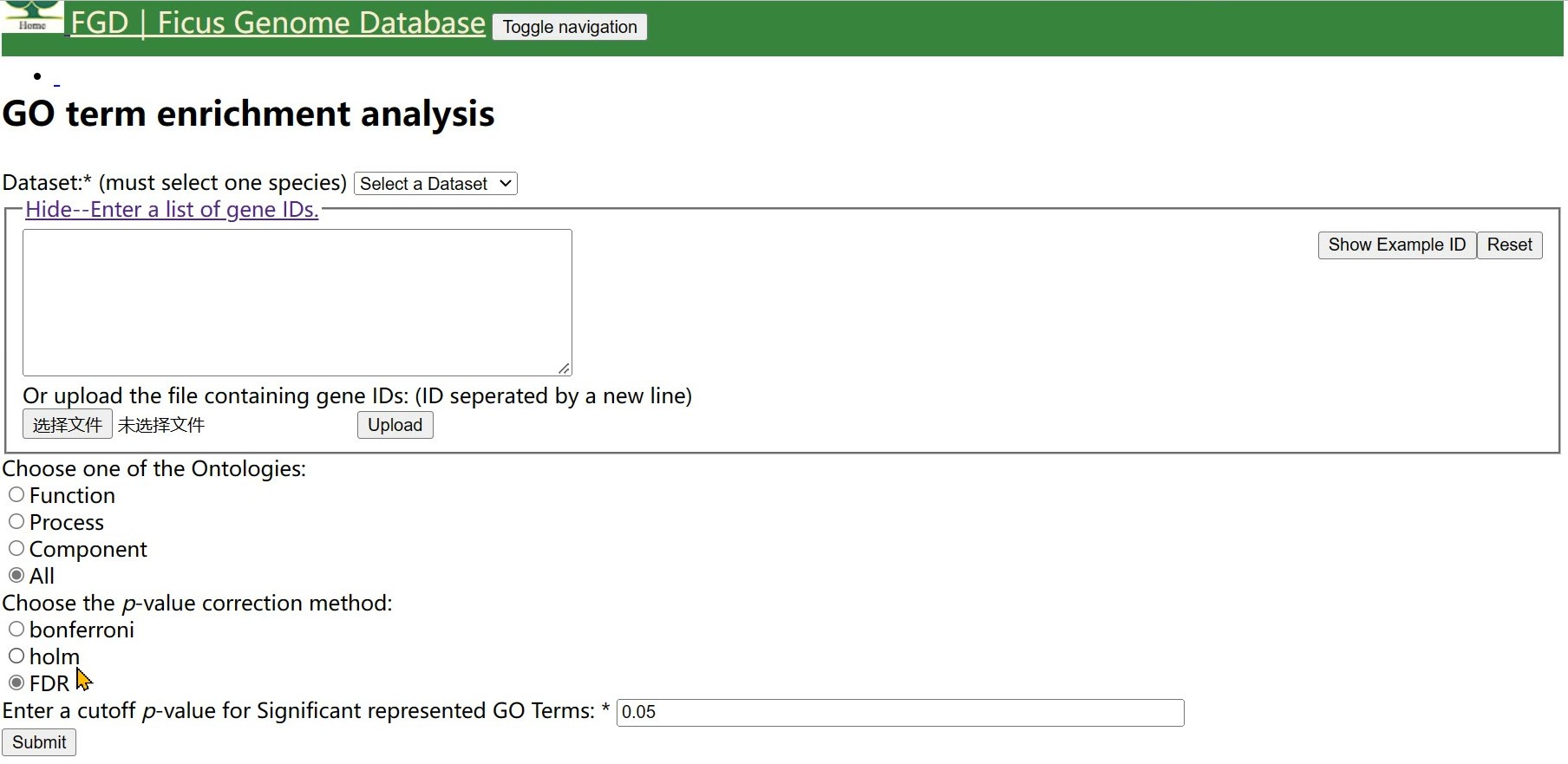
Finally, click on 'submit'.


